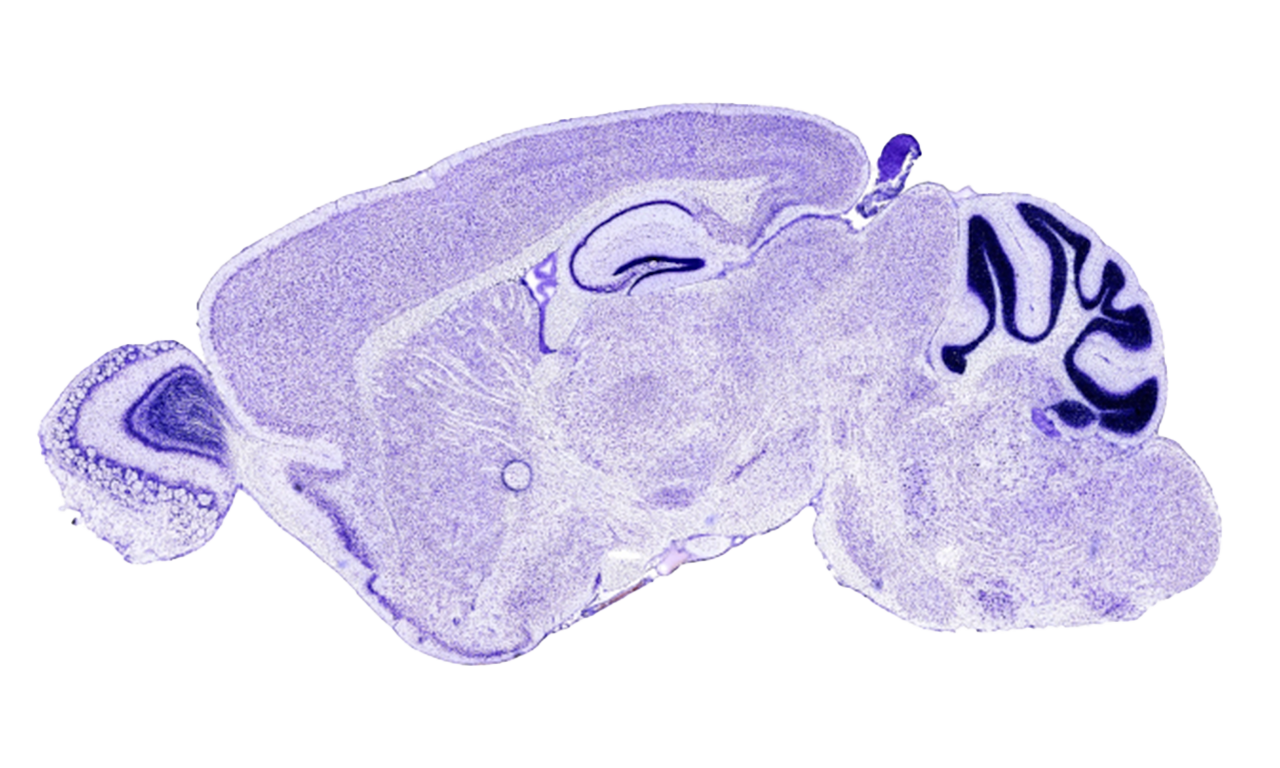
The problem
Image segmentation is the process according to which, starting from an image, the edges of the objects of interest are delimited, isolating them from the background and, therefore, being able to treat them later in an individualized way. In other words, it consists in dividing the image into disjunct regions based on its semantic content. It is an extraordinarily complex and useful task in many real problems. On the one hand, it is a complex task due to the great diversity of existing images: changes in lighting, perspective, deformations of all kinds, intraclass variation of the objects to be segmented, occlusions, changes in size, noise, etc. On the other hand, it is a useful task in innumerable cases: from the segmentation of clavicles in radiographs for the forensic identification of missing people by means of comparative radiography, to the segmentation of the hippocampus in histological images to study gene expression, or the segmentation of anatomical brain structures to follow up the evolution of certain pathologies, just to give three examples.
Methods employed
In the case of the segmentation of mouse brain histological images, deformable models were used (a classic segmentation technique that employs a model whose boundaries expand and contract to fit the contour of the structure to be segmented). The position and evolution of these deformable models were controlled by means of evolutionary computation techniques (particle swarm optimization, differential evolution, real-coded genetic algorithms, and scatter search were tested, and compared with gradient-based approaches), and finally the result was refined using ensemble classifiers (random forest).
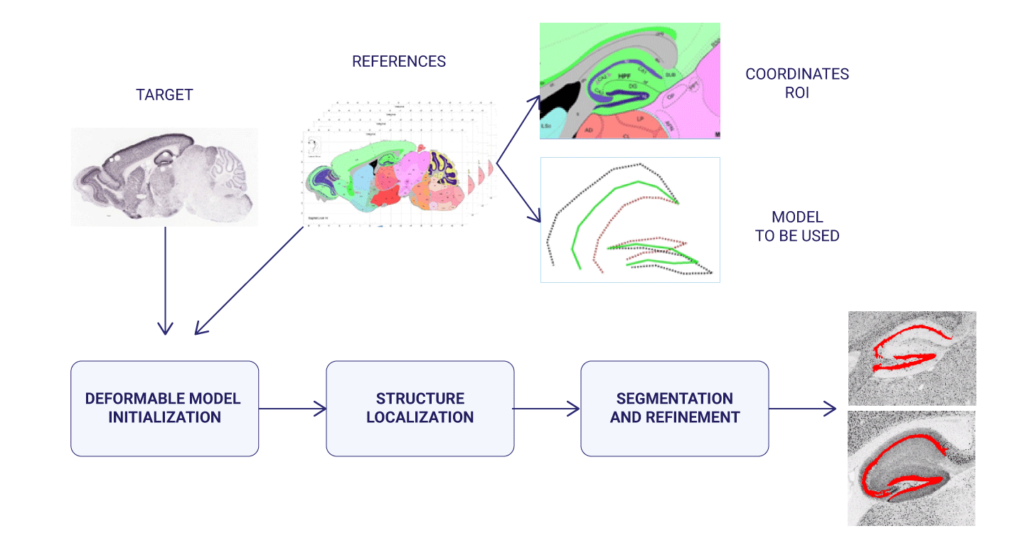
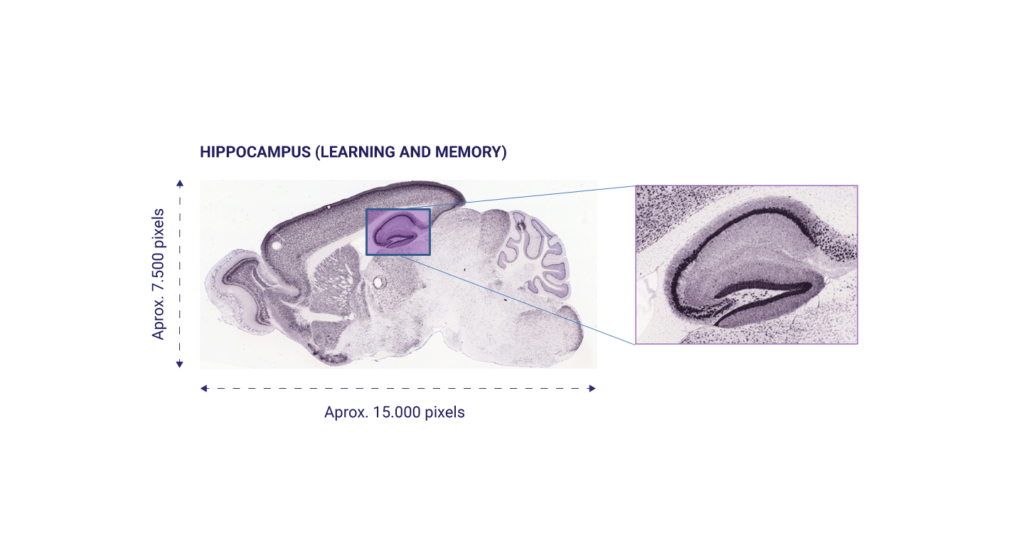
The Allen Mouse Brain Atlas (http://mouse.brain-map.org/) was used, a public database with histological images of the brain in high resolution. This database contains the expression patterns of about 20,000 genes in the mouse adult brain obtained through In Situ Hybridization. This database allows us to study in which part of the brain each gene is expressed, presenting a spatial map of the gene expression patterns of almost every mouse genes (trying to bridging the gap between genomics and neuroanatomy). In particular, the hippocampus was studied, due to its importance in processes such as memory and learning.
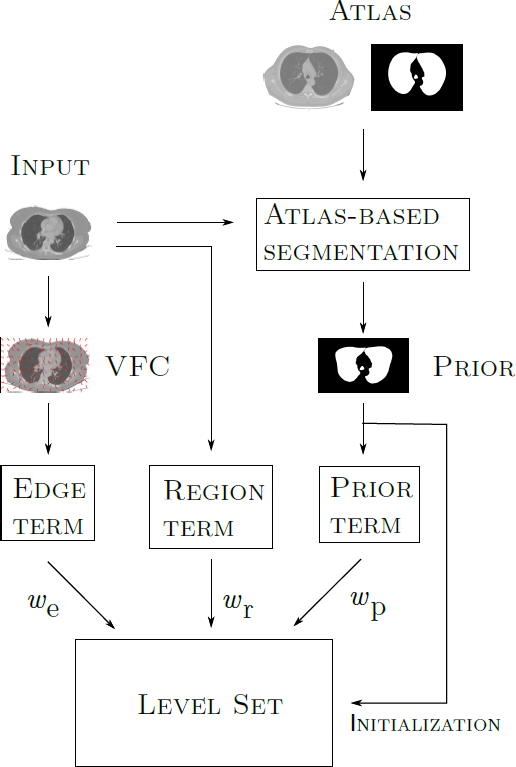
In a later development, in order to solve the same problem, as well as the segmentation of human knees and lungs in Computed Tomography (CT), and brain structures in T1-weighted MRI (caudate, putamen, globus pallidus, and thalamus), a more sophisticated version of the previous method was used, in which deformable models (geometric deformable models, in this case) were initialized through a deformable registration process guided by an evolutionary computation technique; later, the evolution of the deformable model (the Level Set) was determined by region, edge and prior information.
Results
We designed, implemented and validated a system able to automatically locate the hippocampus in mouse brain histological images[1]. This system was applied to the analysis of tens of thousands of high resolution images contained in the Allen Mouse Brain Atlas[2]. This tool allowed to automate and carry out in a few hours a process of gene expression analysis that, if carried out manually, would have taken months. The aforementioned hippocampus localization and segmentation system was also employed in human pose estimation in video sequences[3], and was further generalized to be able to satisfactorily confront other medical imaging modalities and anatomical structures (caudate, putamen, globus pallidus, and thalamus in magnetic resonance imaging, and knees and lungs in CTs)[4]. We therefore provided a medical image segmentation method that was, at the same time, accurate and general. Hybridizations of computer vision (deformable models) and computational intelligence (metaheuristics) techniques were extensively studied and documented[5]. At the time, each of the developments represented an improvement with respect to the state of the art, that is, it improved the results of pre-existing methods. The software developed is available online: https://hal.inria.fr/hal-01221316v3/file/segmentation_framework.zip (Hybrid Level Set Segmentation Framework), https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0074481 (Hippocampus Localization in Brain Histological Images), and http://ibislab.ce.unipr.it/software/libcudaoptimize/doxygen/ (Bio-Inspired Global Optimization Toolbox).
Participants
Institutions
This project was developed at the University of Parma (Italy), in collaboration with the European Center for Soft Computing (Spain), as part of the MIBISOC (“Medical Image using Bio-Inspired Techniques and Soft Computing”) Marie Curie Initial Training Network. MIBISOC was funded by the European Commission with 3.464.246,58€, and involved 8 European research institutions.
Researchers
Dr. Pablo Mesejo as predoctoral researcher at the University of Parma, as part of MIBISOC. Dr. Andrea Valsecchi and Dr. Óscar Ibáñez as predoctoral and postdoctoral researchers at the European Center for Soft Computing, respectively, were also involved in certain parts of this project.
References
[1] Mesejo, P., Ugolotti, R., Di Cunto, F., Giacobini, M., and Cagnoni, S., “Automatic Hippocampus Localization in Histological Images using Differential Evolution-Based Deformable Models”, Pattern Recognition Letters 34 (3), 299-307, Elsevier, February – 2013
[2] Ugolotti, R.*, Mesejo, P.*, Zongaro, S., Bardoni, B., Berto, G., Bianchi, F., Molineris, I., Giacobini, M., Cagnoni, S., and Di Cunto, F., ”Visual search of neuropil-enriched RNAs from brain in situ hybridization data through the image analysis pipeline Hippo-ATESC”, PLoS ONE 8 (9): e74481.doi:10.1371/journal.pone.0074481, Public Library of Science, September – 2013 *These authors contributed equally to this work.
[3] Ugolotti, R., Nashed, Y.S.G., Mesejo, P., Ivekovič, S., Mussi, L., and Cagnoni, S., “Particle Swarm Optimization and Differential Evolution for Model-based Object Detection”, Applied Soft Computing 13 (6), 3092-3105, Elsevier, June – 2013
[4] Mesejo, P., Valsecchi, A., Marrakchi-Kacem, L., Cagnoni, S., and Damas, S., “Biomedical Image Segmentation using Geometric Deformable Models and Metaheuristics”, Computerized Medical Imaging and Graphics 43, 167-178, Elsevier, July – 2015
[5] Mesejo, P., Ibáñez, O., Cordón, O., & Cagnoni, S. (2016). A survey on image segmentation using metaheuristic-based deformable models: state of the art and critical analysis. Applied Soft Computing, 44, 1-29.